In an earlier post I showed four different techniques that enables two-way analysis of variance (ANOVA) using Python. In this post we are going to learn how to do two-way ANOVA for independent measures using Python.
An important advantage of the two-way ANOVA is that it is more efficient compared to the one-way. There are two assignable sources of variation – supp and dose in our example – and this helps to reduce error variation thereby making this design more efficient. Two-way ANOVA (factorial) can be used to, for instance, compare the means of populations that are different in two ways. It can also be used to analyse the mean responses in an experiment with two factors. Unlike One-Way ANOVA, it enables us to test the effect of two factors at the same time. One can also test for independence of the factors provided there are more than one observation in each cell. The only restriction is that the number of observations in each cell has to be equal (there is no such restriction in case of one-way ANOVA).
We discussed linear models earlier – and ANOVA is indeed a kind of linear model – the difference being that ANOVA is where you have discrete factors whose effect on a continuous (variable) result you want to understand.
Python 2-way ANOVA
import pandas as pd
from statsmodels.formula.api import ols
from statsmodels.stats.anova import anova_lm
from statsmodels.graphics.factorplots import interaction_plot
import matplotlib.pyplot as plt
from scipy import stats
In the code above we import all the needed Python libraries and methods for doing the two first methods using Python (calculation with Python and using Statsmodels). In the last, and third, method for doing python ANOVA we are going to use Pyvttbl. As in the previous post on one-way ANOVA using Python we will use a set of data that is available in R but can be downloaded here: TootGrowth Data. Pandas is used to create a dataframe that is easy to manipulate.
datafile="ToothGrowth.csv"
data = pd.read_csv(datafile)
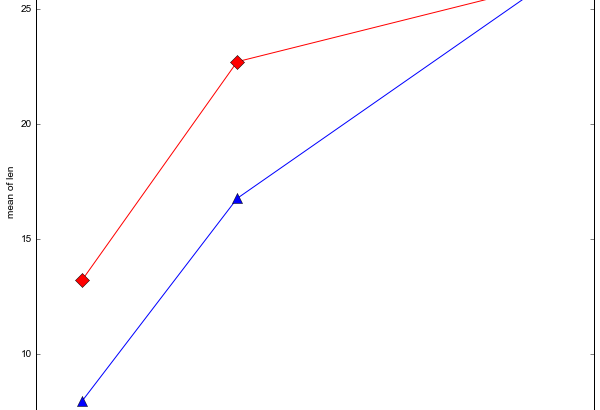
It can be good to explore data before continuing with the inferential statistics. statsmodels has methods for visualising factorial data. We are going to use the method interaction_plot.
fig = interaction_plot(data.dose, data.supp, data.len,
colors=['red','blue'], markers=['D','^'], ms=10)

Calculation of Sum of Squares
The calculations of the sum of squares (the variance in the data) is quite simple using Python. First we start with getting the sample size (N) and the degree of freedoms needed. We will use them later to calculate the mean square. After we have the degree of freedom we continue with calculation of the sum of squares.
Degrees of Freedom
N = len(data.len)
df_a = len(data.supp.unique()) - 1
df_b = len(data.dose.unique()) - 1
df_axb = df_a*df_b
df_w = N - (len(data.supp.unique())*len(data.dose.unique()))
Sum of Squares
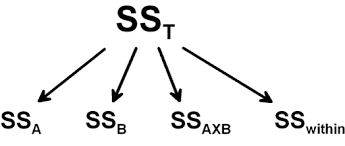
For the calculation of the sum of squares A, B and Total we will need to have the grand mean. Using Pandas DataFrame method mean on the dependent variable only will give us the grand mean:
grand_mean = data['len'].mean()The grand mean is simply the mean of all scores of len.
Sum of Squares A – supp
We start with calculation of Sum of Squares for the factor A (supp).
ssq_a = sum([(data[data.supp ==l].len.mean()-grand_mean)**2 for l in data.supp])
Sum of Squares B – dose
Calculation of the second Sum of Square, B (dose), is pretty much the same but over the levels of that factor.
ssq_b = sum([(data[data.dose ==l].len.mean()-grand_mean)**2 for l in data.dose])
Sum of Squares Total
ssq_t = sum((data.len - grand_mean)**2)
Sum of Squares Within (error/residual)
Finally, we need to calculate the Sum of Squares Within which is sometimes referred to as error or residual.
vc = data[data.supp == 'VC']
oj = data[data.supp == 'OJ']
vc_dose_means = [vc[vc.dose == d].len.mean() for d in vc.dose]
oj_dose_means = [oj[oj.dose == d].len.mean() for d in oj.dose]
ssq_w = sum((oj.len - oj_dose_means)**2) +sum((vc.len - vc_dose_means)**2)
Sum of Squares interaction
Since we have a two-way design we need to calculate the Sum of Squares for the interaction of A and B.
ssq_axb = ssq_t-ssq_a-ssq_b-ssq_w
Mean Squares
We continue with the calculation of the mean square for each factor, the interaction of the factors, and within.
Mean Square B
ms_a = ssq_a/df_a
Mean Square B
ms_b = ssq_b/df_b
Mean Square AxB
ms_axb = ssq_axb/df_axb
Mean Square Within/Error/Residual
ms_w = ssq_w/df_w
F-ratio
The F-statistic is simply the mean square for each effect and the interaction divided by the mean square for within (error/residual).
f_a = ms_a/ms_w
f_b = ms_b/ms_w
f_axb = ms_axb/ms_w
Obtaining p-values
We can use the scipy.stats method f.sf to check if our obtained F-ratios is above the critical value. Doing that we need to use our F-value for each effect and interaction as well as the degrees of freedom for them, and the degree of freedom within.
p_a = stats.f.sf(f_a, df_a, df_w)
p_b = stats.f.sf(f_b, df_b, df_w)
p_axb = stats.f.sf(f_axb, df_axb, df_w)The results are, right now, stored in a lot of variables. To obtain a morereadable result we can create a DataFrame that will contain our ANOVA table.
results = {'sum_sq':[ssq_a, ssq_b, ssq_axb, ssq_w],
'df':[df_a, df_b, df_axb, df_w],
'F':[f_a, f_b, f_axb, 'NaN'],
'PR(>F)':[p_a, p_b, p_axb, 'NaN']}
columns=['sum_sq', 'df', 'F', 'PR(>F)']
aov_table1 = pd.DataFrame(results, columns=columns,
index=['supp', 'dose',
'supp:dose', 'Residual'])
As a Psychologist most of the journals we publish in requires to report effect sizes. Common software, such as, SPSS have eta squared as output. However, eta squared is an overestimation of the effect. To get a less biased effect size measure we can use omega squared. The following two functions adds eta squared and omega squared to the above DataFrame that contains the ANOVA table.
def eta_squared(aov):
aov['eta_sq'] = 'NaN'
aov['eta_sq'] = aov[:-1]['sum_sq']/sum(aov['sum_sq'])
return aov
def omega_squared(aov):
mse = aov['sum_sq'][-1]/aov['df'][-1]
aov['omega_sq'] = 'NaN'
aov['omega_sq'] = (aov[:-1]['sum_sq']-(aov[:-1]['df']*mse))/(sum(aov['sum_sq'])+mse)
return aov
eta_squared(aov_table1)
omega_squared(aov_table1)
print(aov_table1)
Output ANOVA table
| sum_sq | df | F | PR(>F) | eta_sq | omega_sq | |
|---|---|---|---|---|---|---|
| supp | 205.350000 | 1 | 15.572 | 0.000231183 | 0.059484 | 0.055452 |
| dose | 2426.434333 | 2 | 92 | 0.000231183 | 0.702864 | 0.692579 |
| supp:dose | 108.319000 | 2 | 4.10699 | 0.0218603 | 0.031377 | 0.023647 |
| Residual | 712.106000 | 54 |
Two-way ANOVA using Statsmodels
There is, of course, a much easier way to do Two-way ANOVA with Python. We can use Statsmodels which have a similar model notation as many R-packages (e.g., lm). We start with formulation of the model:
formula = 'len ~ C(supp) + C(dose) + C(supp):C(dose)'
model = ols(formula, data).fit()
aov_table = anova_lm(model, typ=2)
Statsmodels does not calculate effect sizes for us. My functions above can, again, be used and will add omega and eta squared effect sizes to the ANOVA table. Actually, I created these two functions to enable calculation of omega and eta squared effect sizes on the output of Statsmodels anova_lm method.
eta_squared(aov_table)
omega_squared(aov_table)
print(aov_table)
Output ANOVA table
| sum_sq | df | F | PR(>F) | eta_sq | omega_sq | |
|---|---|---|---|---|---|---|
| C(supp) | 205.350000 | 1 | 15.571979 | 2.311828e-04 | 0.059484 | 0.055452 |
| C(dose) | 2426.434333 | 2 | 91.999965 | 4.046291e-18 | 0.702864 | 0.692579 |
| C(supp):C(dose) | 108.319000 | 2 | 4.106991 | 2.186027e-02 | 0.031377 | 0.023647 |
| Residual | 712.106000 | 54 |
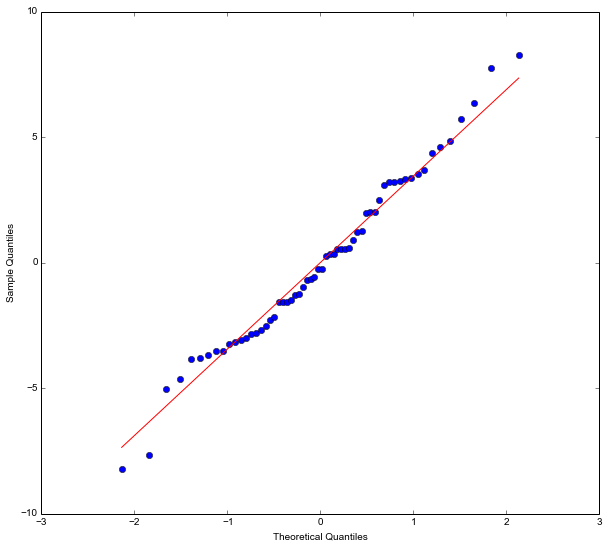
What is neat with using statsmodels is that we can also do some diagnostics. It is, for instance, very easy to take our model fit (the linear model fitted with the OLS method) and get a Quantile-Quantile (QQplot):
res = model.resid
fig = sm.qqplot(res, line='s')
plt.show()
Two-way ANOVA in Python using pyvttbl
The third way to do Python ANOVA is using the library pyvttbl. Pyvttbl has its own method (also called DataFrame) to create data frames.
from pyvttbl import DataFrame
df=DataFrame()
df.read_tbl(datafile)
df['id'] = xrange(len(df['len']))
print(df.anova('len', sub='id', bfactors=['supp', 'dose']))
The ANOVA tables of Pyvttbl contains a lot of more information compared to that of statsmodels. Actually, Pyvttbl output contains an effect size measure; the generalized omega squared.
Measure: len
| Source | Type III Sum of Squares | df | MS | F | Sig. | η2G | Obs. | SE of x̄ | ±95% CI | λ | Obs. Power |
| supp | 205.350 | 1.000 | 205.350 | 15.572 | 0.000 | 0.224 | 30.000 | 0.678 | 1.329 | 8.651 | 0.823 |
| dose | 2426.434 | 2.000 | 1213.217 | 92.000 | 0.000 | 0.773 | 20.000 | 0.831 | 1.628 | 68.148 | 1.000 |
| supp * dose | 108.319 | 2.000 | 54.159 | 4.107 | 0.022 | 0.132 | 10.000 | 1.175 | 2.302 | 1.521 | 0.173 |
| Error | 712.106 | 54.000 | 13.187 | ||||||||
| Total | 3452.209 | 59.000 |
The post Three ways to do a two-way ANOVA with Python appeared first on Erik Marsja.